The MetaFluidics Work package structure follows the workflow of metagenomics (WP2-5) and the whole project is driven by industrial targets with a clear focus on innovation and exploitation of project outputs (WP6).
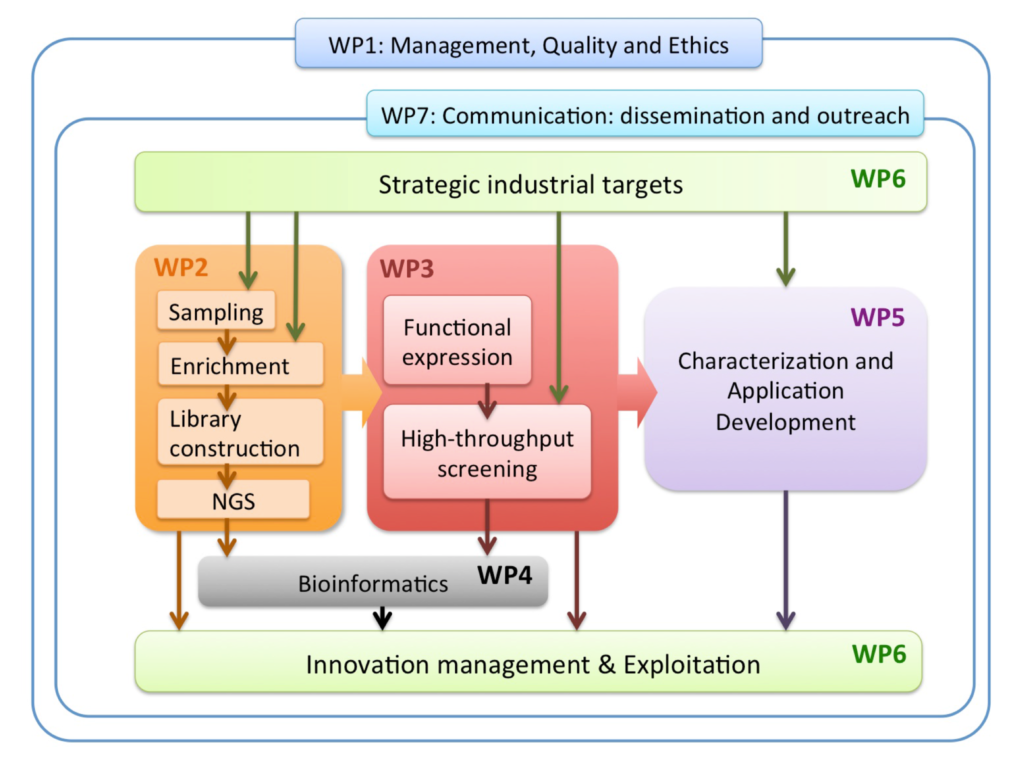
Objectives of the different Work Packages
WP 1: Management (Lead: UAM)
Coordination of overall research tasks to ensure an objective-driven project supervision, EC and H2020 guidelines, with quality and ethics
WP2: Standardization, Sampling and Library Construction (Lead: BIRD)
Procure metagenomic DNA from existing samples and new (complementary) sampling sites, Develop enrichment methods for metagenomes in genetic traits of special interest, construct libraries in suitable expression systems
WP3: Expression and Screening (Lead: UCAM)
Establish extremophillic hosts for metagenome library expression in conventional or microfluidic format.
WP4: Bioinformatics (Lead: QIAAR)
Develop user-friendly and resource-efficient software for metagenomic data analysis
WP5: Application Development (Lead: PROZO)
Analyze the sequences of the recovered hits and characterize functionally and structurally the discovered products.
WP6: Innovation and Exploitation (Lead: DT)
Creation of the plan for dissemination and exploitation of results (PDER) and re-evaluation during project lifetime
WP7: Dissemination, Communication and Outreach (Lead: INSAT)
Create awareness about the project and the Framework program through efficient, targeted communication
WP8: Ethics (Lead: UAM)
set out the ‘ethics requirements’ that the project must comply with.



